Lab database and compute ecosystem
Full stack development of an intergrated lab database and compute system that allows users to easily log/query experimental data and provides tools for performing automated, standardized data analysis pipelines in a high performance computing environment.
Introduction
Reproducible, standardized data analysis pipelines are critically important in research. As the volume and size of the datasets collected grows (as is currently the case in systems neuroscience) this need becomes more apparent. In this project, I developed an internal lab database and compute ecosystem that allows experimental data to be easily logged and queried by users. It also provides the framework and tools to facilitate automated, standardized data analysis using high performance computing by all lab members.
Our lab has developed several custom data processing pipelines that are critical for supporting our everyday research. In the past, team members were expected to run these pipelines on our local lab servers. This approach had two main drawbacks. First, due to limited internal compute resources, these pipelines could often take anywhere from three weeks to over a month to complete, per dataset. Given that we regularly collect 10-15 datasets per week, this timeline was not feasible for our research. Second, the pipelines were usually run in Jupyter notebooks using non-version controlled source code. Therefore, discrepancies between processed data were difficult to track down. With my new system, a single dataset can be processed in 2-4 days and the processing is done with standardized, version controlled source code that is shared between lab members.
This is a large project that I have been independently developing over roughly 3 years in the RoLi Lab. Although I am still working on incremental improvements, the system is fully operational and currently being used by all lab members. Because it lives on our institute’s private network and contains proprietary data, I am not able to go into full details or release any of the associated code or tools I have built here. Therefore, the goal of this post is to provide an overview of the system and its component parts, hopefully illustrating the advantages of building such a system and motivating others to adopt similar ecosystems for data management and analysis in their own research.
Outline
- Overview
- Database
- Web API
- Julia API
- Automated db watchdog
- Max Planck High Performance Computing System
Overview
At a high level, the system is composed of 4 parts:
- Database backend
- Frontend database APIs
- Database watchdog to monitor database entries, send compute jobs, and collects results
- Compute environment (remote HPC system)
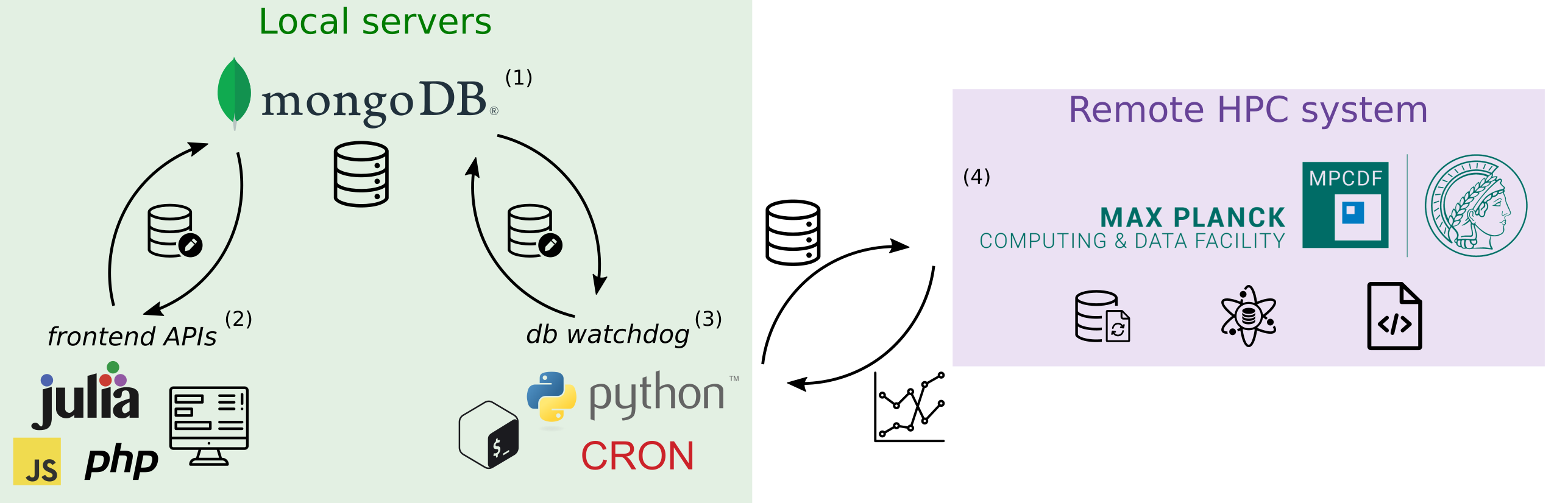
In the following sections, I will unpack each of these components in more detail.
Database
I chose to implement the database backend using a NoSQL approach. Specifically, I used MongoDB. There were two reasons for this.
- NoSQL allows a high level of flexibility in terms of the database schema. This is useful for us, as the types of experiments that are performed in the lab will evolve over time. Thus, a system that can flexibly adapt to log new types of information was desired.
- MongoDB provides nice tools for Replication which ensures that even if one of our lab servers goes down, the database remains live and accessible.
Database collections
The database consists of three separate collections:
- users
- Stores general lab member user information such as email, lab username, HPC account information, and login credentials for the web API
- data
- Stores meta data for every experiment.
- Consists of required fields (e.g., timestamp of data acquisition and location of the data)
- … as well as non-required fields (e.g., free text comments about the experiment)
- queue
- Stores data analyses to be performed on HPC system
- Can be uniquely linked to the
datacollection via the dataset acquisition timestamp
Web API
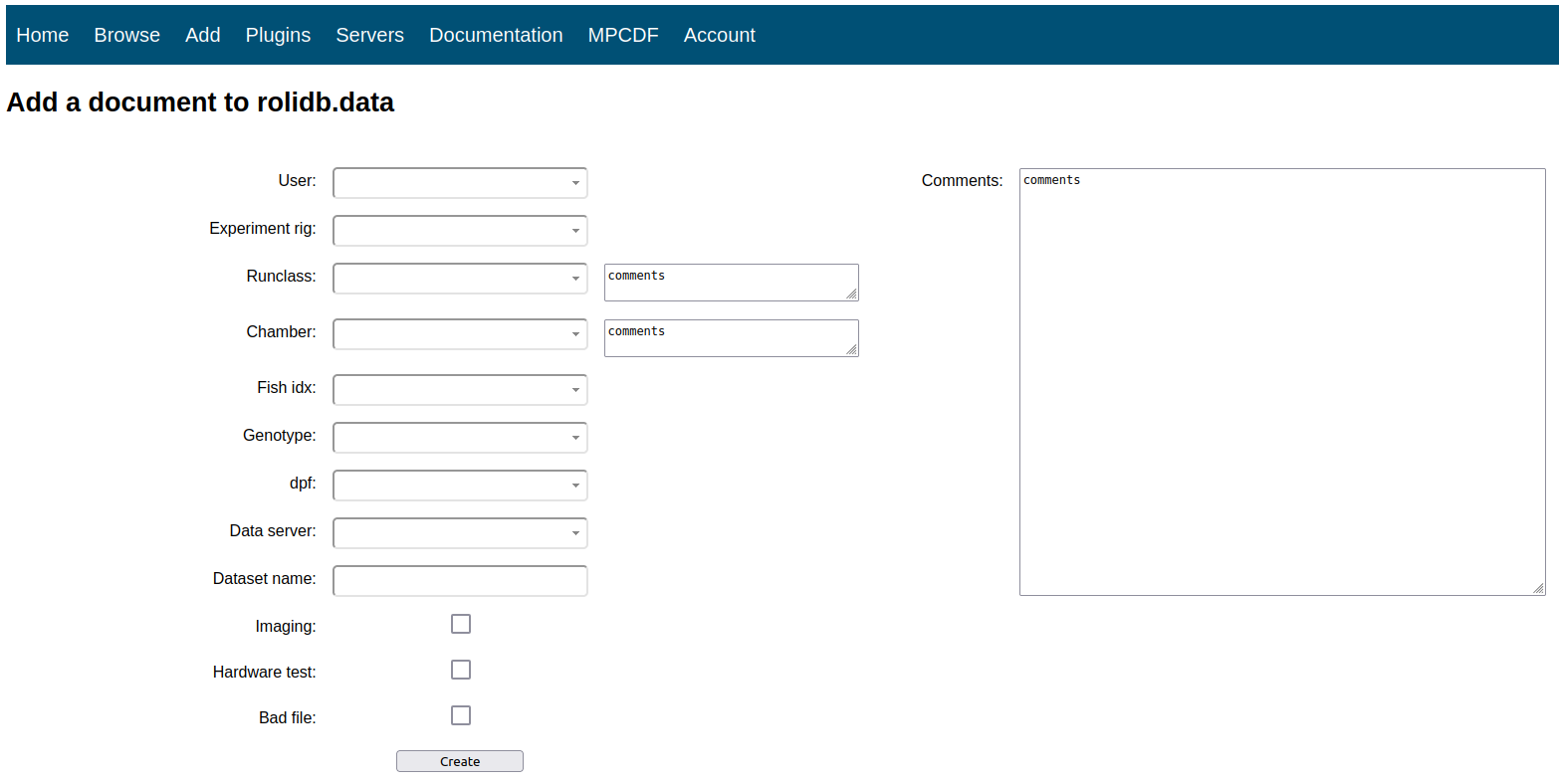
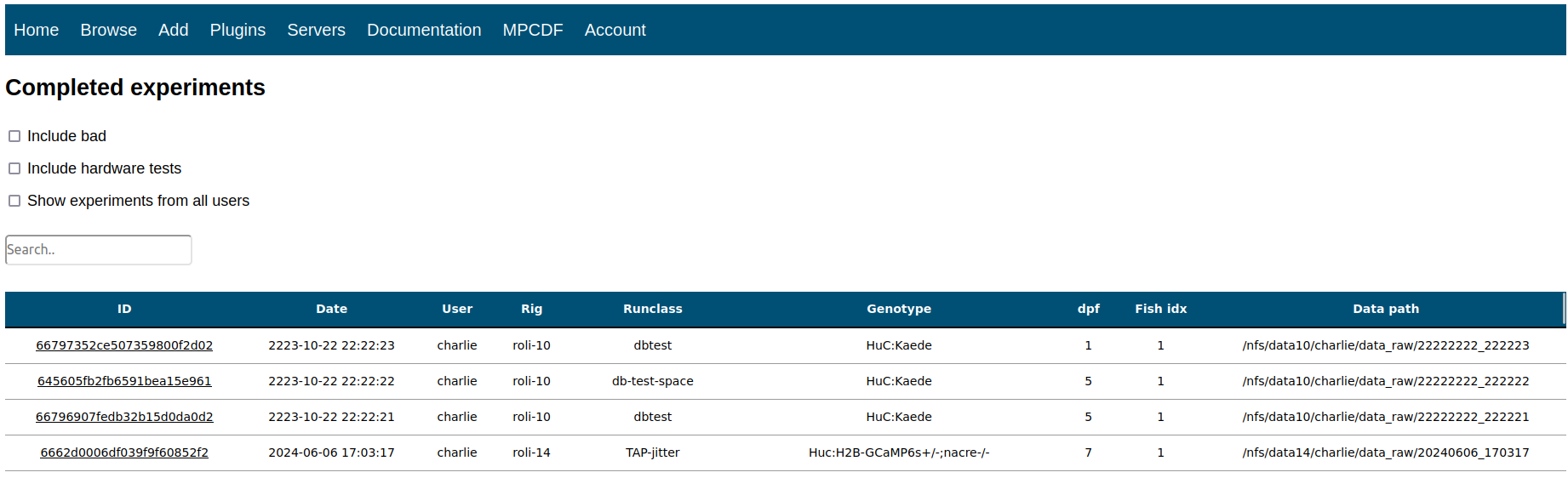
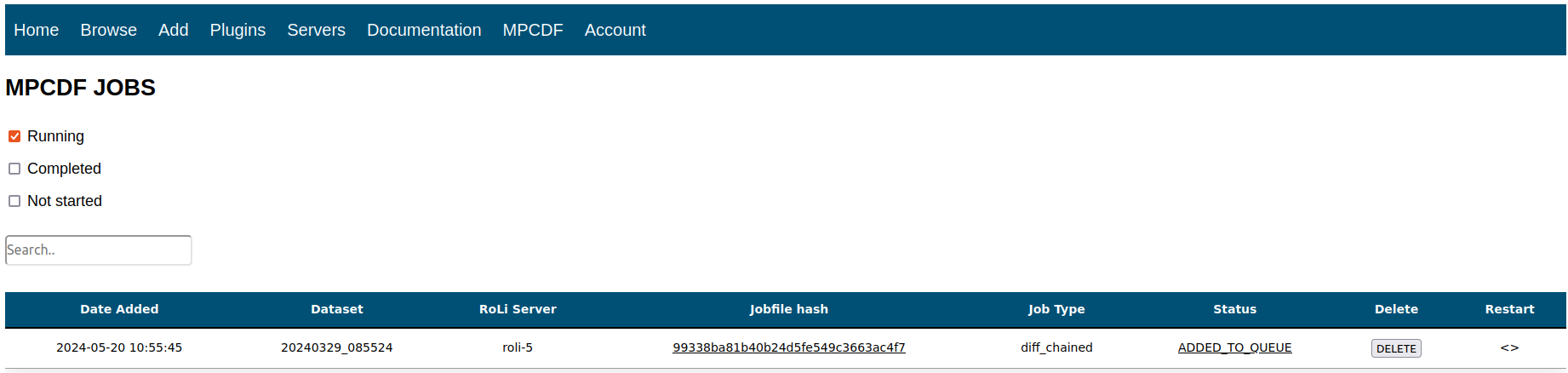
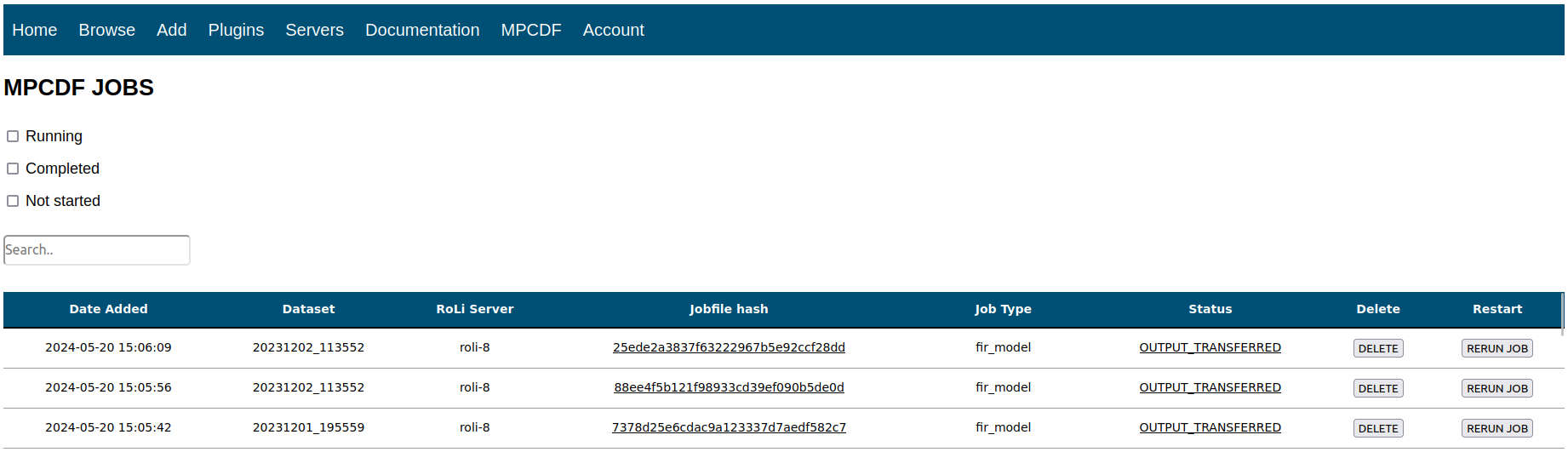
In order to provide a user-friendly interface with the database, I built a lightweight web API which lives in a docker container on a lab server and was built using a combination of PHP, CSS, HTML, and JavaScript. This platform serves 3 main purposes:
- Allow users to enter new experimental data into the database
- Allow user to browse experimental data
- Allow users to monitor HPC job status
Julia API
Our lab performs most data analysis and visualization using Julia. Therefore, I built a very simple Julia API to allow users to interact with the database from code. The two main use cases are:
- Query the database to find a set of datasets to analyze:
julia> dbquery(["data_raw_path", "runclass.id"], filter=Dict("user"=>"charlie", "runclass.id"=>"ARB-diff")) 6×2 DataFrame Row │ data_raw_path runclass.id │ Any Any ─────┼──────────────────────────────────────────────── 1 │ /nfs/data6/charlie/data_raw/2022… ARB-diff 2 │ /nfs/data6/charlie/data_raw/2022… ARB-diff 3 │ /nfs/data6/charlie/data_raw/2022… ARB-diff 4 │ /nfs/data6/charlie/data_raw/2022… ARB-diff 5 │ /nfs/data7/charlie/data_raw/2022… ARB-diff 6 │ /nfs/data1/charlie/data_raw/2022… ARB-diff
- Submit batch jobs to the HPC system
julia> dbqueue_job( dataset="20211215_093527", user="charlie", compute_cluster="raven", job_type="diff_registration", data_transfer_await=false, mpcdf=true, datafiles=["phase_and_reference.h5", "registration_preps.h5", "reconstruction_ref_sweep.h5"], datarawfiles=["fl_a.roli", "fl_b.roli"], job_options=Dict("split_idx" => 1, "num_splits" => 40) )The above command creates a new entry in the
queuecollection which will get scraped by the db watchdog system that I discuss in the next section.
Automated queue monitoring
Running a data analysis pipeline on a High Performance Computing (HPC) system has huge advantages in terms of speed, scalability, and reproducibility. However, getting it set up can sometimes be challenging, particularly for those without prior experience writing batch scripts or working with job scheduling systems such as SLURM. In order to run a data analysis job on an HPC system, you need to transfer your data to where it can be accessed by the compute nodes, build a “batch script” (the set of instructions that tells the machine what to do with the data), submit this batch script to SLURM, and you need to locate the output of your job and move it back to a local server.
To streamline this process for standard data pipelines in our lab, I built an automated system that allows users to easily utilize our institute’s HPC system. As a result of this automation, from the perspective of the user, all that needs to be done to submit a job is run one line of code in Julia, monitor the job’s status on the web API, and wait for the results to be returned to their local machine in lab.
This automation is achieved by a set of CronJobs running on our local lab servers. These CronJobs contain 3 steps:
- Monitor the queue collection to find new entries
- Python script to scrape the queue collection in the lab database and look for new job entries
- When a new job is found, initiate the transfer of the data to the remote compute node using
rsync - After the data is transferred, transfer the job batch script to the compute node
- When a new job batch script is detected on the compute node, automatically add to SLURM queue
- Monitor the status of in progress transfers
- Python script to query the queue collection and find jobs with status = transfer in progress
- Quantify transfer progress by capturing
rsync --progressoutput and adding this information to the database
- Monitor the status of in progress jobs
- Capture SLURM PID upon being added to job queue
- Monitor remote results directory for analysis output file
- When final output is detected, initiate transfer of output file(s) back to local lab server
All of these tools live in a single git repository. In order to get started using the system, users just need to clone the repo and run a short python install script which sets up the user’s cron tab, requests the user’s credentials, and mounts the remote data directory (using sshfs) in order to achieve automation. In addition to these automation tools, this repository also contains a folder with the lab analysis source code to be executed on the remote HPC system. This ensures that all users analyze their data using the same procedure which increases the standardization and reproducibility of data pipelines in our lab.
Max Planck High Performance Computing System
All Max Planck Institute researchers have access to state-of-the-art HPC systems. These are managed by the MPCDF. Further details can be found by visiting their website. For the project described here, we have primarily made use of the raven compute cluster, however, my system was built to be able to utilize additional clusters and I am in the process of incporating the new viper cluster, as well.